The membrane that encases a biological cell is not simply a barrier; it is chock full of proteins involved in all sorts of critical biological functions. To really understand what membrane proteins are doing and how, researchers need to know how they’re organized and how they interact with one another. But uncovering that information is challenging.
Yale researchers have now developed a new microscopy method called Native-nanoBleach that overcomes the main challenges to understanding membrane protein organization, including the difficulty in studying these membranes without disrupting the native environment and limits to the resolution of light microscopes typically used to study them.
And to demonstrate the effectiveness of the new method, they successfully applied it to a biological conundrum—pertaining to proteins involved in the development of pancreatic cancers and how they might be targeted for treatment—that has remained unsolved for decades.
They describe the new method and its advantages in a new study published in Nature Nanotechnology.
Methods typically used in the study of membrane protein organization require removing the native membrane environment surrounding proteins and then putting isolated proteins of interest into environments that mimic but do not fully replicate the complexity of the real cell membrane, said Moitrayee Bhattacharyya, assistant professor of pharmacology at Yale School of Medicine and senior author of the study. This approach, Bhattacharyya said, removes important context since the proteins interact with the molecules that surround them.
Second, light microscopes, which are commonly used to observe protein organization, don’t have the resolution needed to determine whether proteins near each other are truly interacting or are simply neighbors on membranes.
Last, the amount of a particular protein found in a cell membrane may be too low or too high for current methods of study. In those instances, researchers have to make adjustments; they either have to replicate proteins that in their natural state are too few in number or separate out proteins from samples where there are too many. But this again can remove important context about the natural state of proteins as they sit and function in cell membranes.
“Ideally, we would have a method that would work with any endogenous level of cell membrane protein expression,” said Bhattacharyya.
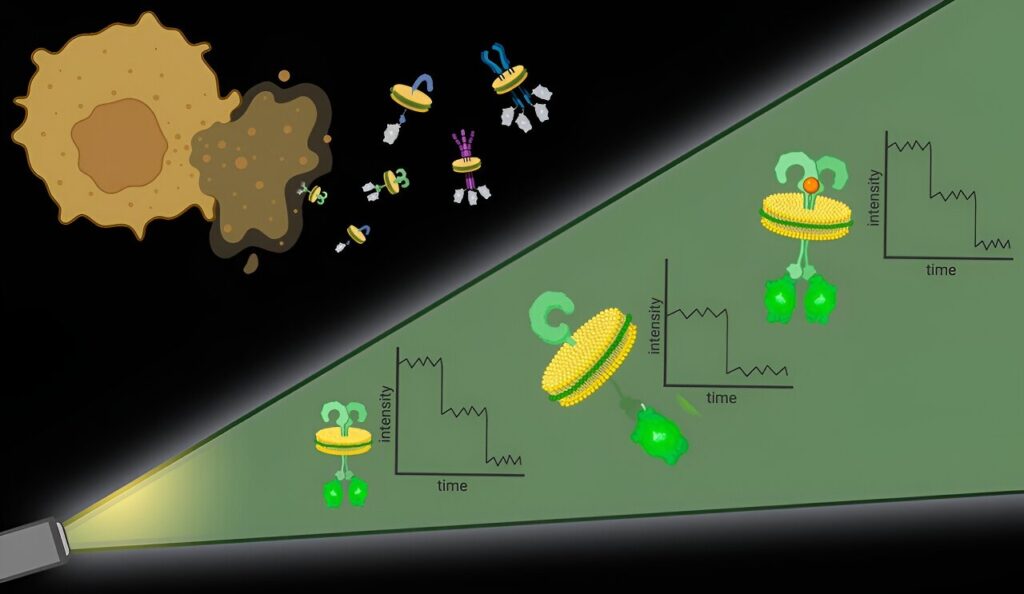
To tackle the first challenge, the researchers used particular molecules, types of polymers, to essentially punch out protein complexes with their surrounding cell membrane intact. “It’s like the cell membrane is a sheet of cookie dough and the polymers are cookie cutters,” said Bhattacharyya.
These bits of protein with surrounding cell membrane, dubbed native nanodiscs, are approximately 10 nanometers in diameter, small enough that any proteins contained in the nanodisc are likely interacting, which addresses the second challenge. Further, this approach works with any cell membrane protein in any amount, allowing researchers to observe proteins at their natural levels in native membranes.
Once the nanodiscs are generated, researchers can use any number of commonly used techniques to hone in on a particular protein of interest. They then quantify the proteins in each nanodisc with the help of fluorescent molecules attached to them.
It’s an approach that offers high spatial resolution without the need for specialized hardware, said Bhattacharyya.
“This work presents a new technique to understand how membrane proteins—which represent about 60% of drug targets—assemble into functional units on or within the native lipid bilayer,” said Gerard Walker, co-first author of the paper and a graduate student in Bhattacharyya’s lab.
To demonstrate how this method might be applied, the researchers took on a decades-long debate in biology. A protein called KRas is mutated in more than 90% of human pancreatic cancers, sparking immense clinical and therapeutic interest. Whether KRas subunits come together to form dimers (two units) or oligomers (more than two units) on cell membranes has remained the focus of longstanding investigation.
Studies, however, have produced conflicting findings. Animal and cellular studies, which lack detailed molecular resolution, show evidence that KRas units come together on cell membranes. Meanwhile, biophysical analyses, which do not retain the native membrane around proteins, have found KRas remains in single units, or monomers.
“With our method, we get the best of both worlds,” said Bhattacharyya. “We retain the native membrane environment and we have very high spatial and single-molecule resolution. When we applied our method, we found that KRas exists as dimers and monomers in similar amounts. But when KRas is mutated, as in pancreatic cancers, dimers increase and monomers decrease.”
The finding highlights the importance of the native cell membrane for understanding membrane proteins and identifies a target—reducing KRas dimerization—for cancer treatment. This is just one of many ways this method could be used to understand the role of membrane protein organization in disease, Bhattacharyya said.
“It’s really rewarding to see Native-nanoBleach already being successfully applied to a wide variety of pressing biological questions in the Bhattacharyya lab and beyond,” said Caroline Brown, co-first author of the study and a Ph.D. candidate in the lab of co-author Kallol Gupta, assistant professor of cell biology.
Membrane proteins make up a third of all of the proteins in the human body and this approach can be used to study any of them, said Bhattacharyya.
“It’s a general technique,” she said. “There’s really no limitation.”
Going forward, Bhattacharyya and her colleagues hope to extend this approach to studying protein organization in the membranes of various organelles, structures, like mitochondria, that are contained within cells.