A research team led by the late Professor Liang Haojun from the Hefei National Laboratory for Physical Sciences at the Microscale of University of Science and Technology of China (USTC) has developed a facile enthalpy-mediated strategy to precisely control the replication and catalytic assembly of DNA-functionalized colloids in a time-dependent manner, facilitating the creation of large-scale ordered nanomaterials. The study was published in Angewandte Chemie International Edition.
The replication of information is a fundamental characteristic of nature, with nucleic acids playing a crucial role in biological systems. However, creating synthetic systems that can produce large-scale, three-dimensionally ordered nanomaterials using self-replicating nanostructures has remained a formidable challenge.
Existing artificial self-replicating systems often fall short in programmable assembly into sophisticated nanostructures, limiting their potential functions and applications.
The research team devised an innovative solution to overcome the existing challenges. By leveraging the power of DNA’s specificity and the principles of dynamic DNA nanotechnology, they have established a template system that includes DNA-functionalized colloidal seeds.
These seeds were paired with a simplified DNA strand-displacement circuit programmed subsystem, which was designed to produce DNA-functionalized colloidal copies. This system operated at room temperature, eliminating the need for complex and potentially damaging high-temperature processes.
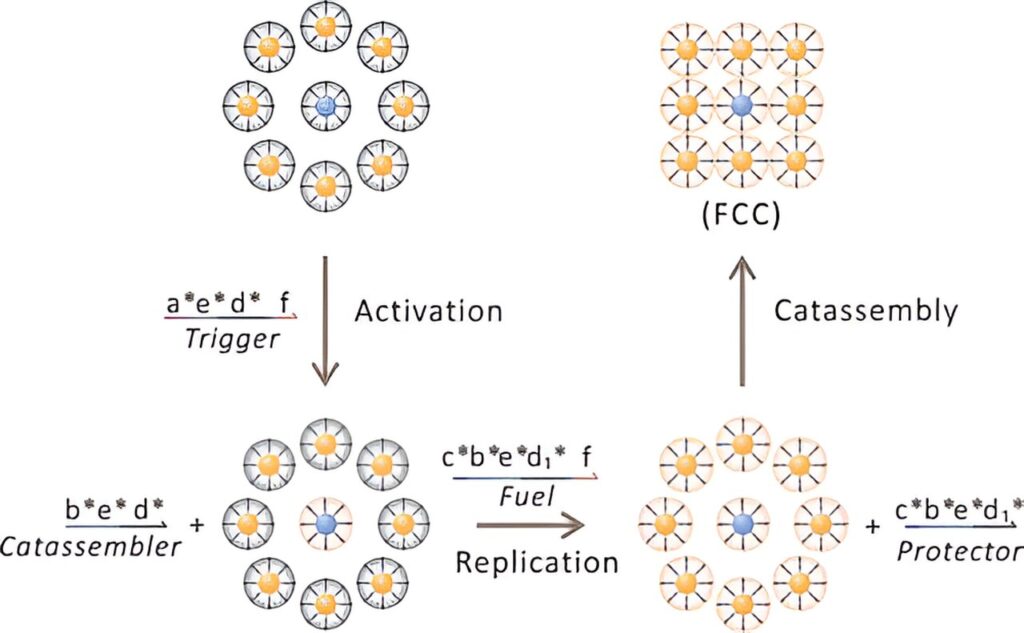
The key innovation lied in the use of a time-dependent interaction system that controlled the replication and catalytic assembly of programmable atom equivalents (PAEs). The PAEs, consisting of colloidal cores functionalized with DNA, were capable of recognizing and binding complementary DNA strands through a process known as toehold-mediated strand displacement (TMSD) reactions.
This allowed for the controlled release of catalysts from the template system, which in turn initiated the replication of PAE seeds and the assembly of PAE copies into superlattice structures.
Through programmed DNA assembly, inactive PAE copies gradually acquired the same sticky ends as PAE seeds, facilitating replication and subsequent assembly into ordered superlattices. Importantly, the system demonstrated remarkable accuracy in recognizing and transmitting template information during the replication process, ensuring fidelity in the reproduction of superlattice structures.
The study opens avenues for the construction of complex, programmable, large-scale three-dimensional colloidal superlattice materials, which could find applications in fields ranging from materials science to biotechnology.
Provided by
University of Science and Technology of China