Northwestern Medicine investigators led by Ruli Gao, Ph.D., assistant professor of Biochemistry and Molecular Genetics, have developed a novel genetic sequencing tool that accelerates sequencing analysis of same-cell genotypes and phenotypes in tumors, as detailed in a study published in Nature Communications.
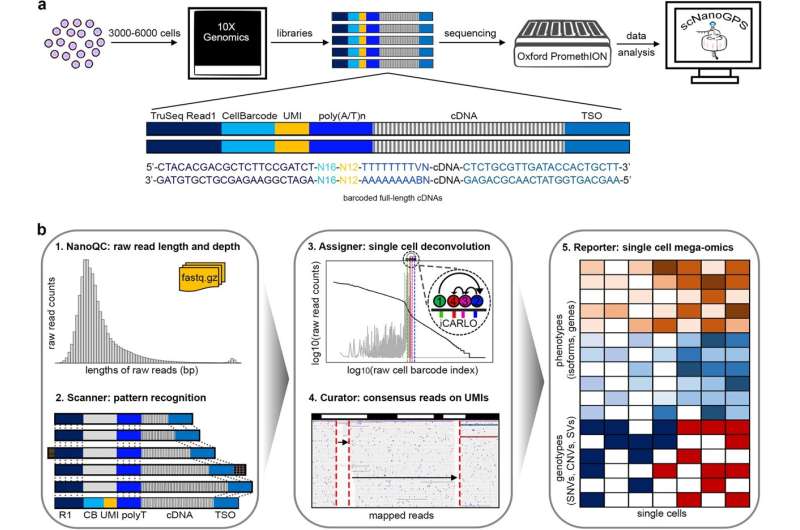
Single-cell nanopore RNA sequencing is a newer type of genetic sequencing that pushes forward current high-throughput single-cell RNA sequencing techniques from next-generation sequencing (NGS), which can only sequence short strands of RNAs, to long-read third-generation sequencing (TGS), which can directly measure the full length of RNAs.
However, the advanced technique is also known to produce high sequencing errors and also relies either on short-reads sequencing—generating matched NGS data to guide the identification of cellular data—or on using a barcode whitelist to split the data into true cells and single molecules.
“This technique relies on cell barcodes and single molecule barcodes, what we call unique molecule identifiers, to achieve high throughput, so you see the challenges due to higher sequencing errors in barcode sequences. Currently available methods depend on next-generation data or depends on a theoretical whitelist to recognize which are true cell barcodes and unique molecule identifiers,” said Gao, who is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
To improve this process, Gao’s team developed scNanoGPS (single-cell Nanopore sequencing analysis of Genotypes and Phenotypes Simultaneously), a new sequencing tool that performs completely independent deconvolution of error-prone long-reads into single cells and single molecules.
“To further facilitate the applications of novel technology in genomics research, scNanoGPS constructs additional functional modules to calculate both genotypes (mutations) and phenotypes (gene and isoform expressions) in individual cells from high-throughput single-cell nanopore RNA sequencing data,” said Cheng-Kai Shiau, Ph.D., a postdoctoral fellow in the Gao laboratory and co-first author of the study.
scNanoGPS increases sequencing throughput from hundreds to thousands of cells, which is comparable to current widely applied short-reads based single-cell RNA sequencing techniques, and the expression profiles between short-read and long-read data are highly concordant, according to Gao.
“Due to whole gene body coverages, long-read data enables calculations of splicing isoforms and genetic alterations that are largely missed by short-read single-cell RNA sequencing,” Gao said.
To validate their tool, Gao’s team used scNanoGPS to sequence kidney tumor cells and lymphocytes, finding that the cells express cell-type specific combinations of isoforms and genetic mutations.
“We found cell-type specific isoforms including both tumor cell specific isoforms, and immune cell specific isoforms, and also demonstrated cell-type specific mutation profiles within the same tumors,” said Lina Lu, Ph.D., a postdoctoral fellow in the Gao laboratory and co-first author of the study.
“It’s known that alternative splicing isoforms is a critical post-transcription mechanism to increase protein complexity in human cells. We are excited to note that scNanoGPS enables the direct measurement of splicing isoforms at single-cell levels,” Gao said.
As for future applications, Gao hopes scNanoGPS can be used to identify cell-type specific isoforms that contribute to diverse human diseases such as cancer, heart failure and even organ transplantation rejection.
“Novel technologies make data generation relatively easier, however researchers cannot use new data types effectively without a robust computational tool. scNanoGPS fills a gap between the technologies and applications. We hope to make the powerful long-read single-cell sequencing technology available and accessible to all general labs,” Gao said.