In plants and animals, the basic packaging units of DNA, which carry genetic information, are the so-called nucleosomes. A nucleosome consists of a segment of DNA wound around eight proteins known as histones.
During gene expression (the process lying at the basis of protein production), nucleosomes are involved in various dynamical structural changes, such as nucleosome sliding, DNA unwrapping and other DNA–histone interactions. Of particular importance in these processes are the end structures, or tails, of the histones.
Histone tails undergo chemical modifications, changing the histone’s functionality as needed. Detailed studies, and especially visualizations, of nucleosome dynamics are crucial for better understanding the role of histone tails.
Mikihiro Shibata from Kanazawa University and colleagues have now succeeded in making video recordings of tail-less nucleosomes, showing that the absence of histone tails significantly increases a nucleosome’s dynamic activity. The study is published in Nano Letters.
The scientists used high-speed atomic force microscopy (HS-AFM), a powerful nanoimaging tool for visualizing molecular structures and their dynamics at high spatial and temporal resolution.
For this, the nucleosomes needed to be put onto a substrate. Shibata and colleagues used a film of so-called pillar[5]arenes (molecules with a pentagonal tubular structure) as the substrate, forming an ideal surface as the nucleosomes are easily adsorbed to it without dynamical processes getting suppressed.
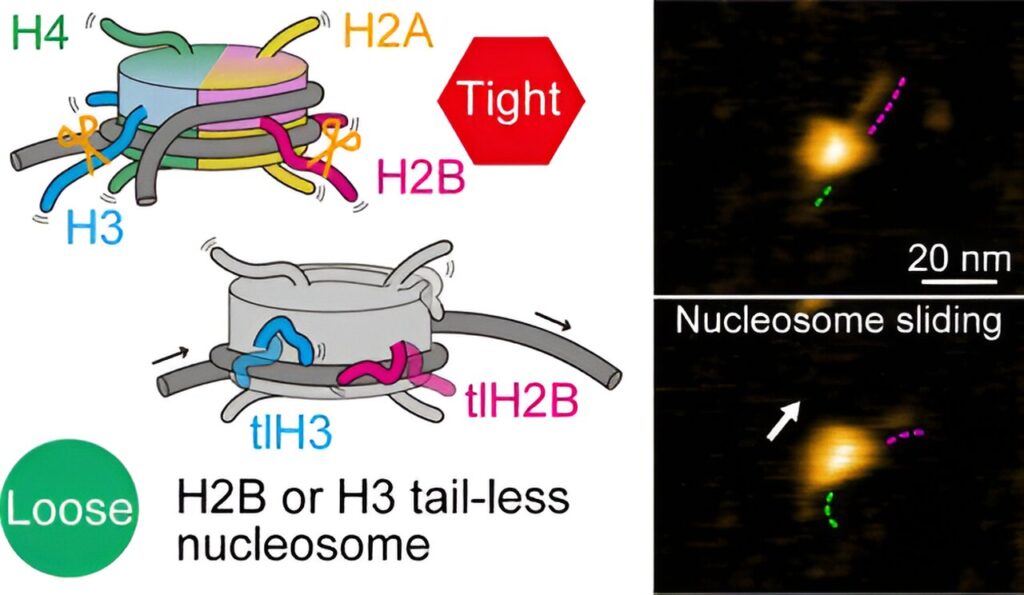
The researchers first looked at nucleosomes for which all eight histones lacked tails. Based on their HS-AFM observations, they concluded that nucleosome sliding and DNA unwrapping/rewrapping occurred more often than for normal (canonical) nucleosomes. This suggests that without tails, the histone–DNA interaction is weakened, leading to a situation in which DNA can more easily detach from the histones.
To better understand the roles of specific histone tails, Shibata and colleagues prepared nucleosomes where one type of histone was tailless. There are four different types of histones, called H2A, H2B, H3 and H4. HS-AFM experiments on the nucleosomes revealed that H2B and H3 tail-less nucleosomes showed an increased frequency of dynamics. Conversely, this means that canonical H2B and H3 histones are essential for nucleosome stability.
The scientists point out that they could not observe any actual motion of histone tails—most likely the temporal resolution of the study, 0.3 seconds, was much slower than the rate of the wrapping/unwrapping dynamics of the tails. Despite this limitation, the work of Shibata and colleagues clearly proves that the tails of H2B and H3 histones are the main contributors to nucleosome dynamics.
Regarding future work, quoting the researchers, “a technique for tagging histone tail tips might enable HS-AFM to capture the movements of the histone tails themselves.”
High-speed atomic force microscopy
The general principle of atomic force microscopy (AFM) is to make a very small tip scan the surface of a sample. During this horizontal (xy) scan, the tip, which is attached to a small cantilever, follows the sample’s vertical (z) profile, inducing a force on the cantilever that can be measured.
The magnitude of the force at the xy position can be related to the z value; the xyz data generated during a scan then result in a height map providing structural information about the investigated sample. In high-speed-AFM (HS-AFM), the working principle is slightly more involved: the cantilever is made to oscillate near its resonance frequency.
When the tip is moved around a surface, the variations in the amplitude (or the frequency) of the cantilever’s oscillation—resulting from the tip’s interaction with the sample’s surface—are recorded, as these provide a measure for the local z value. AFM does not involve lenses, so its resolution is not restricted by the so-called diffraction limit as in X-ray diffraction, for example.
HS-AFM results in a video, where the time interval between frames depends on the speed with which a single image can be generated (by xy-scanning the sample).
Researchers at Nano Life Science Institute (WPI-NanoLSI), Kanazawa University have in recent years developed HS-AFM further, so that it can be applied to study biochemical molecules and biomolecular processes in real-time. Mikihiro Shibata and colleagues have now applied the method to study nucleosome dynamics in detail, and in particular the role of the molecular endings of histones—proteins that play a crucial role in DNA accessibility.