Generally speaking, the higher the degree of information restoration of a vaccine to a virus, the greater its potential efficacy. The virus itself is the most authentic vaccine, such as the varicella-zoster virus, which provides lifelong immunity after a single infection. However, viruses also evolve mechanisms to evade immune surveillance during their long evolutionary history, such as evading the immune system’s pursuit by frequently changing disguises through high mutability.
Alternatively, they may lower their own visibility and lurk invasively through special mechanisms, and coronaviruses are adept at employing both of these tactics.
As an RNA virus, coronaviruses have a natural advantage in high mutability. Meanwhile, named ‘corona’ due to the crown-like protrusions on their surface, coronaviruses display the most crucial antigenic information on the receptor-binding domain (RBD) protein located at the top of these corona-like protrusions.
The antigenic information is scattered among the solitary peaks on the viral surface, resembling a corona. This spatially discrete structure is challenging for the immune system to recognize effectively.
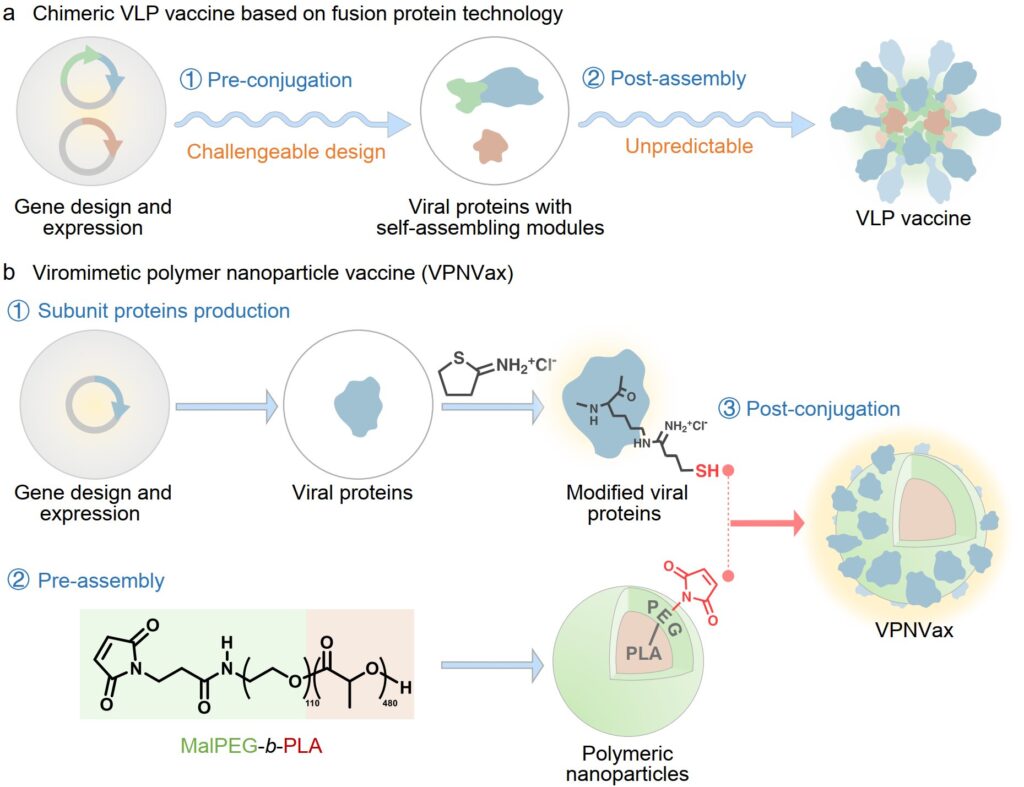
Addressing the structural characteristics of coronaviruses, a team led by Professors Xuesi Chen and Wantong Song from the Changchun Institute of Applied Chemistry reported a viromimetic polymer nanoparticle vaccine (VPNVax). The vaccine was prepared by rearranging the RBD proteins of the coronavirus and modifying them onto the surface of pre-assembled polyethylene glycol-polylactic acid polymer nanoparticles.
This modular preparation strategy offers several advantages: (1) it allows for flexible control of antigen density (valence) on the surface of the nanoparticle vaccine; (2) enables the substitution of antigen proteins to respond to outbreaks of different virus variants swiftly; (3) facilitates the direct transformation from subunit proteins to nanoparticle vaccines, streamlining the process of rapid large-scale preparation.
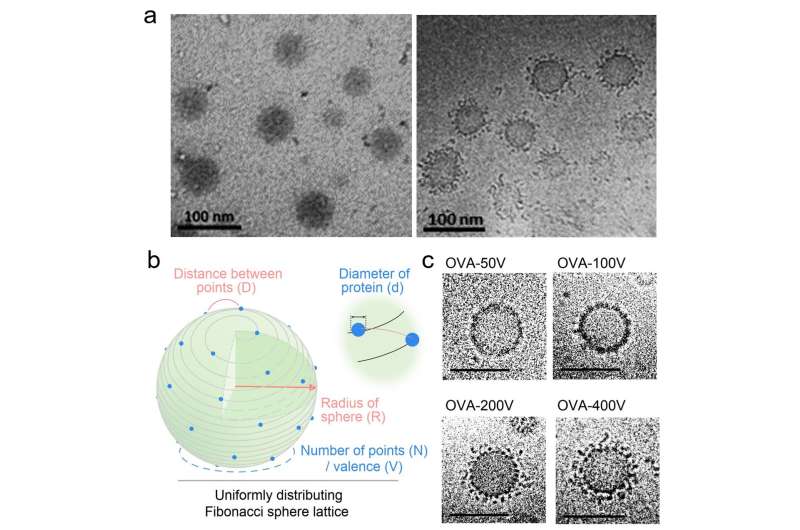
The morphology of VPNVax under cryo-electron microscopy is extremely similar to the virus structure, with antigen proteins densely distributed on the spherical surface of the nanoparticle carrier. Through theoretical calculations using the Fibonacci spherical lattice model and the regulation of chemical reaction conditions, the research team successfully prepared VPNVaxs with different surface valences.
Results showed that the surface antigen valence indeed had a significant impact on the immune stimulatory effect of the nanoparticle vaccine. A higher antigen density on the surface of VPNVax enhances its direct activation capability on B cells, indirectly validating the coronavirus’s mechanism of evading immune surveillance by reducing surface antigen density through corona-like protrusions.
This also underscored the necessity of optimizing and controlling the surface valence of nanoparticle vaccines. However, excessively high valency also reduced the structural stability of VPNVax, necessitating a moderate valence to achieve a balance between stimulatory effects and stability.
The research team further discovered that for antigen proteins of different sizes, the optimal immune stimulatory effect of the prepared VPNVax occurred when the surface protein coverage was in the range of 20%-25%. Furthermore, the VPNVax with the optimal structural parameters, when combined with commercial aluminum adjuvants, achieved a stronger immune stimulatory effect, and its immune serum had been proven to have virus-neutralizing effects.
More importantly, this polymer-based vaccine platform can further develop and exploit the adjuvant function of the polymer carrier. By carrying immune agonists or regulating the polymer’s chirality, VPNVax could simultaneously activate cellular immune responses.
In summary, the research conducted on the VPNVax platform regarding the structure-effect relationship of nanoparticle vaccines and the preparation strategy that combines materials synthesis technology offers new insights for designing the next generation of virus-like particle vaccines.
The work is published in the journal National Science Review.