Just as the name suggests, DNA origami is a fabrication technique wherein researchers fold DNA strands to create precisely shaped 2- and 3-dimensional nanostructures. These highly programmable structures have the potential to transform drug delivery, diagnostic medicine, nanomaterials formation and molecular computing, but much like crafting paper origami, scientists are limited to the designs they can conceptualize.
To overcome creative blocks, researchers in the Department of Mechanical Engineering at Carnegie Mellon University have developed a generative design tool that can produce optimally-driven wireframe DNA origami nanostructures conforming to designer-defined constraints.
“Scientists can now generate hundreds of nanostructures that fit their specific needs in minutes,” said A.J. Vetturini, mechanical engineering Ph.D. candidate.
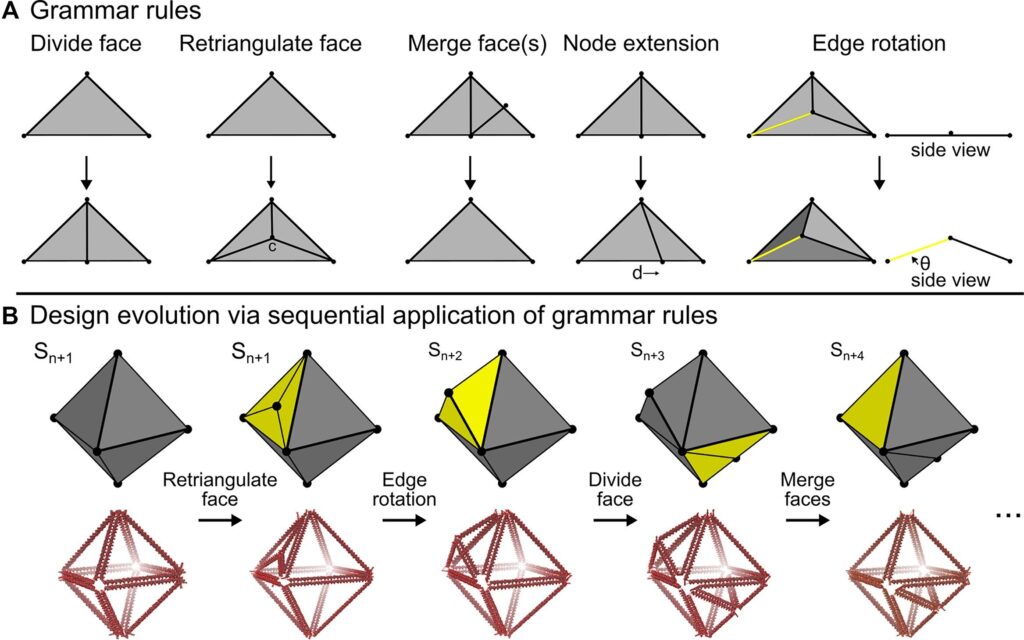
Used in the design of products and architecture, shape grammar rules are planning tools that help manipulate shapes to fit within defined rules to generate a new design. Because DNA’s makeup is determined by a simple set of rules (A pairs with T, C pairs with G), applying shape grammars to DNA origami was an easy choice for the research team, who created such rules for guiding the formation of DNA structures.
The new computer-aided design tool featured in Nucleic Acids Research, relies on grammar rules to cycle through design iterations before outputting the best solution. This generative design process called shape annealing was pioneered by Jon Cagan, professor of mechanical engineering.

“With this tool, we can manufacture structures that people wouldn’t necessarily think of on their own, but now they can exist in the world with a purpose,” he said.
Drawing on the team’s deep knowledge of DNA nanotechnology, they realized that the vast majority of existing designs explore very little of the design space. First, the complexity of creating highly-anisotropic DNA origami shapes often prevents exploration of potentially groundbreaking designs. Further, designs are often highly constrained in terms of available material since most DNA origami have fixed sizes due to their use of standardized and commercially-available single-stranded DNA “scaffolds.”
“Designing nanostructures is extremely difficult because it requires carefully plotting thousands of nucleobases in a specific order,” explained Rebecca Taylor, associate professor of mechanical engineering.
“Achieving a successful design that meets desired outcomes in terms of mechanics, shape and material use is a tremendous challenge without the incorporation of multi-objective optimization in the design process. For that reason, I’m delighted that our newly-released tool is so easy to use and will allow scientists of all levels to generate diverse designs. This could expedite advancements in biomedical and nanotechnology applications.”
Further, the team manufactured and characterized multiple designs produced by this new tool, allowing them to show that this new pipeline is fully compatible with existing approaches to converting 3D meshes into base-level representations of DNA origami.